We are interested in the knowledge and technologies that enhance our ability to model micro-physiological and disease processes in vitro. We apply these knowledge and technologies into diagnostic, therapeutic and toxin testing applications.
Fundamental studies: we focus on the systematic descriptions of the liver regeneration processes at the multi-cellular levels described by mechanical and biochemical networks.
Title: Novel in vitro inflammatory liver model for detection of immune-mediated effects of hepatotoxic compounds
Description: The predictivity of current in vitro liver models remains low because they do not incorporate the important inflammatory effects associated with liver toxicity, as they lack the relevant cell sources. We engineer inflammatory models with co-culture of novel human iPSC-derived Kupffer cells and liver parenchymal cells to recapitulate critical secretory factors associated with inflammation to differentiate immune-mediated effects of hepatotoxic compounds.
Title: Novel In Vitro Liver System for Modeling Nonalcoholic Steatohepatitis (NASH) and aiding in NASH therapeutics
Description: Owing to an increase in the prevalence of obesity, non-alcoholic-fatty-liver disease (NAFLD), a hepatic manifestation of the metabolic syndrome, is now the most common liver disease worldwide. There is currently no approved therapy for NAFLD or NASH, the active form of NAFLD due to a lack of models to study 1) the efficacy/safety of therapeutic compounds and 2) mechanisms of disease progression. The objective of this project is to develop a model which can be used to study NASH and aid in NASH therapeutics. We engineer robust NASH chips with co-culture of iPSC-derived Kupffer cells and liver parenchymal cells to recapitulate critical phenotypic markers for screening compounds affecting NASH progression into liver cancer. Our platform can be used to test efficacy and safety of NASH therapeutics, making it highly attractive for biomedical applications.
Title: 2.5D in vitro hESC model for embryogenesis study and developmental toxicity screening
Description: Human embryogenesis is a complex progress that is delicately controlled by a set of molecular and cellular regulations. However, it is unfeasible obtaining human embryos to carry out comprehensive studies due to the ethical concern. We’d recently developed an in vitro micropatterned-hESCs model that recapitulates gastrulation, and self-organizes to three primary germ layers that the cell fates are relative to the boundary in a fixed distance scale. By utilizing this powerful tool, now we’re able to thoroughly investigate how mechanical and biochemical cues symphonize in the process of human embryogenesis. Furthermore, this in vitro model is also suitable for high-throughput and large-scale developmental toxicity screening, making it a robust tool not only in basic study but also in industrial application.
Title: Cultured pork project
Description: Cell-based meat is meat produced by cultivating animal-derived cells in vitro and using said cells to form desired tissues for consumption. Pork is the most consumed meat in the world. In Asia, demand has risen in recent years and is not expected to slow down any time soon. However, present methods of meat production are unsustainable. Intensive farming of pigs is unethical and poses environmental and health problems. We work on developing sustainable cell sources, scaffolds and culture conditions for constructing textured cuts of pork, with emphasis on pork belly because of its simple layered structure.
Mechanobiology framework for understanding biological function

Integrating mechanical attributes with biochemical molecular events to dissect cellular and tissue level functional processes can yield causal mechanistic insights that are useful for applications. We have studied the contraction process of a liver lumen structure called bile canaliculi that serves the important function of delivery bile from liver to intestine. Such a contraction process is dissected into repeated cycle with each cycle composed of 3 phases. The first phase starts with a rapid expansion driven by bile entry into the lumen, followed by strengthening of the actomyosin wall resisting the lumen expansion causing bleb and vesicle formation often seen in cholestatic diseases prominently. The phase 1 to 2 transition likely is triggered by temporary increase in membrane/cytoskeleton tension that can be measured by a novel confocal phase microscope. Finally massive contraction of the lumen occurs caused by luminal calcium entering the adjacent cells via mechanosensitive calcium channel PIEZO-1. The calcium wave originated from the lumen triggers the local paracellular actomyosin contraction autonomously that likely propagate to neighboring areas to support the directional delivery of the bile down to the bile duct and the intestine. Such an integrated understanding of the bile canaliculi contraction process yield important mechanistic insights that are useful in developing therapeutic or drug testing assays against cholestasis.
Kinectin-dependent ER sliding regulates adhesion maturation
The endoplasmic reticulum (ER) is the largest organelle in the cell, and is a master regulator of diverse cellular functions. The ER is dynamic and extensively spread in the cell; it comes in contact with other organelles and cellular structures to regulate their function. However, the mechanisms of ER transport and signalling have not been fully understood. Integrin-mediated adhesions are one of the cellular structures regulated by the ER. The ER may promote their assembly and maturation. Integrin-mediated adhesions link the extracellular matrix to the actin cytoskeleton, and are involved in diverse physiological and pathological processes such as normal development, immune response and cancer. Our proposed study bring together two important fields of study, ER and integrin-mediated adhesion dynamics. We will address the key questions in the regulation of ER signalling and of integrin-mediated adhesion maturation. The knowledge from this study will contribute to diverse applications such as improvements on stem cell differentiation, the design of biomaterials and therapies for cancer metastasis.
The ER needs to be transported to and mediate contact with adhesion sites to promote adhesion maturation. In our previous AcRF Tier 2 grant, we have shown that this process is mediated by integral ER membrane protein kinectin through microtubule (MT) motor protein kinesin-1. Kinesin-1 mediates ER transport through ER sliding mechanism, but does not bind directly to ER. Despite ER sliding being the preferential mode of ER movement, the main interaction partner of kinesin-1 for ER sliding has not been established. Although kinectin interacts with kinesin-1, it is unknown whether kinectin is the linker between kinesin-1 and ER during ER sliding on acetylated MTs. Acetylation of MTs promotes adhesion maturation, but how this occurs is not understood. Although kinesin-1-dependent ER sliding preferably occurs on acetylated MTs, this mechanism has not been explored because kinesin-1 was previously believed to mediate adhesion disassembly. Since we have shown that kinesin-1 can mediate adhesion maturation through kinectin, we hypothesize that MT acetylation mediates adhesion maturation through kinesin-1/kinectin-dependent ER sliding. The ER may promote adhesion maturation through various signalling pathways, such as through ER-bound proteins PTP1B. The extent of kinesin-1/kinectin-dependent ER sliding in PTP1B signalling to regulate adhesion maturation is not fully understood. Our proposal will determine the role of ER in adhesion maturation by further characterizing the ER transport mechanism, its regulation and downstream signalling. We hypothesize that kinectin mediates kinesin-1-dependent ER sliding on acetylated MTs for transport of PTP1B to support adhesion maturation.
Applications: we develop innovative in vitro cell-based models for hepatotoxicity testing of drugs, pathogens, environmental toxins and agrochemicals; quantitative descriptions of pathological features with optical imaging tools; and develop therapeutics for patients with liver diseases. While staff and students are given resources to explore new ideas, hypotheses and generate supporting preliminary findings, mature concepts are driven to completion by integrated teams; training researchers to adapt to industry environments.
Cultured pork project
Cell-based meat is meat produced by cultivating animal-derived cells in vitro and using said cells to form desired tissues for consumption. Pork is the most consumed meat in the world. In Asia, demand has risen in recent years and is not expected to slow down any time soon. However, present methods of meat production are unsustainable. Intensive farming of pigs is unethical and poses environmental and health problems. We work on developing sustainable cell sources, scaffolds and culture conditions for constructing textured cuts of pork, with emphasis on pork belly because of its simple layered structure.
There are 3 project groups as outlined below:
1. QUANTIFYING 3D epithelial tissue niche to re-define tissue biology and pathology standards at cellular and molecular levels: (development of high throughput and high content optical imaging instruments, animal models of liver and other GI track diseases, and analysis of complex cellular and molecular image features. A translational biomedical research in collaboration with the Institute of Bioengineering and Nanotechnology, or IBN; the Singapore-MIT Alliance for Research and Technology, SMART; and Southern Medical University, China).
Development of AI-based robust digital pathology approaches for liver disease classification
Accurate assessment of liver diseases such as fibrosis and NASH suffer from the data variations of the sampling error, staining and imaging heterogeneity in variable clinical lab settings. Experienced pathologists deal with such variations by comparing with other scoring samples in their memory to generate predictive decisions by selecting useful features and neglecting noises. Accordingly, we have developed machine-/deep-learning algorithms on variable quality and imaging conditions of standard light microscopes, commonly available collagen-stains and variable sizes and quality of biopsy samples. We could accurately detect various stages of liver fibrosis and NASH, even intra-stage cirrhotic changes, and applicable for liver fibrosis caused by different etiologies. Using only conventional collagen and H&E stained liver samples, our approaches makes it possible to quantitatively score liver diseases in fully automated manner and partially correct the sampling error in less sophisticated clinical or laboratory settings and applicable for multi-center collaborations and clinical trials.
2. ENGINEERING 3D micro- tissue building blocks (cellulosic sponges and organoid cultures; hESC/hiPSC-derived human kupffer/hepatic stellate cells; a collaboration with the Institute of Bioengineering and Nanotechnology, or IBN; and Singapore Immunology Network, or SigN).
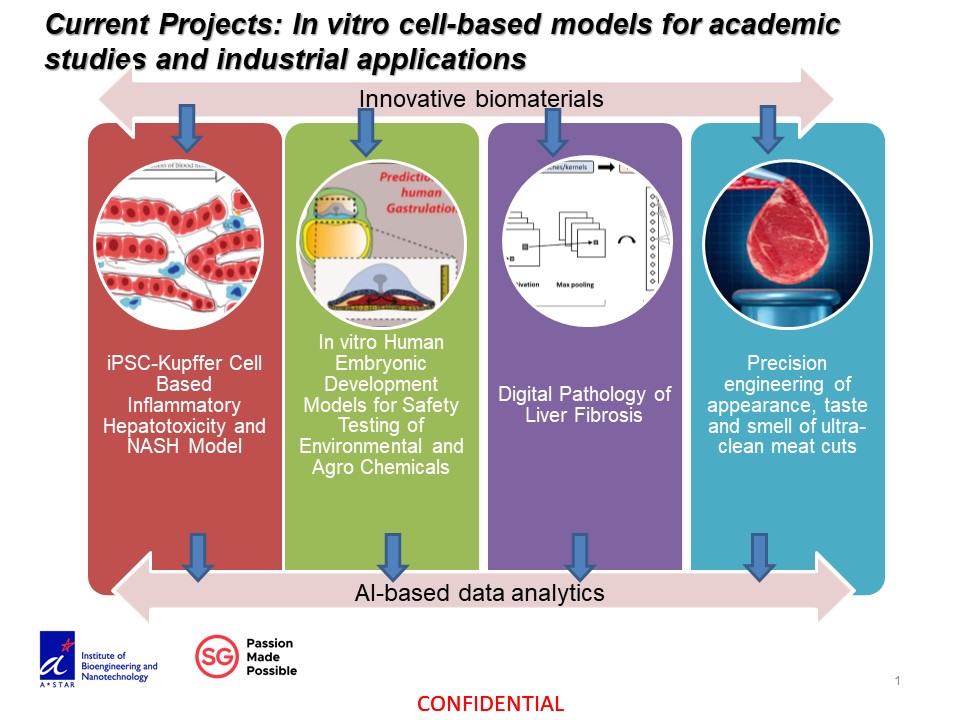
Development of macroporous hydrogel sponges for soft tissue organoid culture and applications
Soft tissue organoids have found various applications in disease modelling and drug testing. Forming organoids is relatively straightforward but maintaining them in culture can be tricky. We had previously developed a class of soft macroporous cellulose sponges for forming and maintenance of liver cell spheroids. We also demonstrated its ability to preserve the heterogeneity and complexity of patient-derived xenograft of liver cancer. To retrieve the spheroids/organoids for applications and characterization, we have developed a variant that is readily dissolved with reducing agent TCEP. A liver progenitor cell differentiate rapidly in the 3D organoid configuration. To enable high-content and higher throughput applications in drug screening, we further developed a transparent variant that form soft macroporous hydrogel sponge directly in multi-well cell culture plates. These developments enable the studies and drug testing applications at different scales.
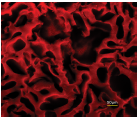
Figure 1. macroporous sponges in aqueous solution
Figure 2. HepaRG differentiation into hepatocytes and cholangiocytes
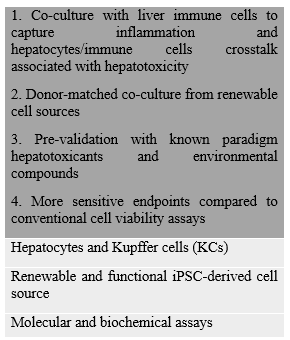
Generation of Mature Kupffer Cells from Human Induced Pluripotent Stem Cells
Liver macrophages, Kupffer cells (KCs), play a critical role in drug-induced liver injury (DILI) and liver diseases including cholestasis, liver fibrosis and viral hepatitis. Yet, studies deploying KCs in in vitro models of DILI and liver diseases are hindered due to variable quality and limited source of human KCs. In vivo, KCs originate from MYB-independent, yolk-sac derived macrophage progenitors, which proliferate and differentiate into liver-specific macrophages in response to hepatic cues in the liver. Here, we recapitulated KCs ontogeny by differentiation of MYB-independent human induced pluripotent stem cells (iPSCs) to macrophage precursors and exposing them to hepatic cues to generate iPSC - derived KCs (iKCs). iKCs were characterized for macrophage - and KC-specific marker expression and functions. iKCs were successfully applied to inflammation-associated in vitro liver model of hepatotoxicity and cholestasis through establishment of iKCs - hepatocyte co-culture and validation of the model using paradigm compounds. iKCs expressed macrophage markers (CD 11, CD14, CD68, CD163 and CD32) at 0.3-5 folds of primary adult human KCs (pKCs). iKCs also expressed KC-specific CLEC-4F, ID1 and ID3, phagocytosed and secreted Interleukin-6 and Tumor Necrosis Factorupon stimulation at levels similar to pKCs but different from non-liver macrophages. iKCs-hepatocyte co-culture model was more sensitive in detecting hepatotoxicity induced by inflammation-associated drugs, Acetaminophen and Trovafloxacin, and cholestasis induced by Chlorpromazine when compared to hepatocytes alone. Hence, iKCs were mature, liver-specific and functional. Furthermore, donor-matched iKCs and iPSC-hepatocyte co-culture exhibited minimal non-specific background response compared to donor-mismatched matched primary hepatocyte-KCs co-culture. iKCs offer a mature renewable human cell source for liver-specific macrophages, useful in developing in vitro model to study DILI and liver disease such as cholestasis. Ongoing work is supported by A*STAR IAF-PP on donor-matched co-culture of hiPSC-derived hepatocytes and kupffer cells.
3. INTEGRATING 3D cell-based assays for testing drugs, environmental toxins, developmental toxins and agrochemicals (in vitro toxicology platforms in collaboration with major industry partners, US Environmental Protection Agency, and the Institute of Bioengineering and Nanotechnology, or IBN).
Ensemble of Multi-disciplinary Systems and Integrated Omics for NAFLD (EMULSION) diagnostic and therapeutic discovery (funded by A*STAR IAF-PP)
Development of Non-Alcoholic Fatty Liver Disease or NASH on chip with patient-derived liver progenitor organoids co-cultured with non-parachymal cells from human sources: a medium throughput robust perfusion 3D culture platform for drug testing.
Application and refinement of predictive in vitro models of hepatotoxicity using environmental and industrial chemicals (funded by A*STAR IAF-PP)
The primary objective of this collaborative project is to further refine predictive models of hepatotoxicity that have been developed using EPA’s ToxCast chemical prioritization program (NCCT) by using rodent liver HTS models being developed at (A*STAR). The predictive models of rodent hepatotoxicity developed by the EPA focused on mining empirical mappings between ToxCast bioactivity assays and adverse chronic hepatic outcomes in rats from ToxRefDB. This research will examine the influence of three main factors with respect to their relevance for improving the predictive accuracy of rodent hepatotoxicity. First, this collaboration will investigate the performance of primary rat hepatocytes in contrast to a mostly HepG2, cell free or non-hepatic assays (currently used in ToxCast Phase). Second, this research will investigate the improvement in predictive accuracy afforded by measuring in vitro bioactivity using sentinel stress response pathways. Third, this research will compare the accuracy for predicting chronic and sub-chronic rat guideline testing outcomes.
NCCT and A*STAR will collaboration on identifying the relevant chemicals, and applying machine learning methods to develop predictive models with the bioactivity data. A key objective of this work is to evaluate the time and concentration-dependent effects of chemicals on sentinel stress response pathways in the rat hepatocytes, and to use these data for predictive modeling. We will use a machine learning approach to objectively compare the performance of the rat HTS assay for classifying rodent hepatotoxicants, and to analyze the gain in sensitivity afforded by this physiologically relevant platform.
Heterogeneity and solutions in cell-based models for in vitro toxicity testing applications
As in vitro cell-based models were developed to better mimic the in vivo toxin responses, complexity and heterogeneity increases that generate large variations that pose challenges to toxicity testing applications. We use two examples to illustrate such challenges and our solutions. One is the environmental toxin testing effort (together with the US EPA) with a few in vitro cell-based models of different complexity. The more biomimicry models unfortunately exhibit inherently large variation and thus low accuracy even though highly functional primary cells together with large data readout and AI-based data analytics on moderately complex model yield the best results. Another example is the developmental toxin testing assay where a model has been developed to properly recapitulate the in vivo developmental process such as cell differentiation and migration. This model again has the inherent heterogeneity which can be tackled with large image-based readout and AI-based data analytics approach. We propose that this approach can be adopted to leverage the plethora of cell-based models made available to eventually generate highly predictive toxicity testing platforms in vitro.